Part I: Antibiotic Resistance (AMR) - Global and Local Epidemiology
1.6 Carbapenem-resistant Enterobacterales (CRE)
-
Enterobacterales can acquire resistance to carbapenem through production of carbapenemase (Table 1.7), modification of outer membrane permeability and efflux pump. [63]
Carbapenemase, KPC-producing K. pneumoniae was first discovered from a clinical isolate through the Intensive Care Antimicrobial Resistance Epidemiology (ICARE) surveillance in North Carolina in 1996 [64,65] and followed by a substantial spread in New York, [66] Israel, [67] and Greece. [68] Enterobacterales producing KPC has also been described in South America (Colombia, Brazil and Argentina) [69–71] and China. [72,73] Other than K. pneumoniae, the KPC-enzyme has also been described in many other Enterobacterales species. [65] Infections caused by carbapenem-resistant organisms increases the risk of complications and mortality. [74]
-
New Delhi metallo-β-lactamase 1 (NDM-1) was first described in 2009 in a Swedish patient of Indian origin. He was hospitalised in India and acquired urinary tract infection caused by a carbapenem-resistant K. pneumoniae. [75] Like other metallo-β-lactamases, the enzyme NDM-1 can hydrolyse all β-lactams except aztreonam. Resistance to aztreonam is usually due to the coexisting ESBL or AmpC β-lactamase. Majority of the NDM-1-producing organisms harbour other resistance mechanisms, rendering them resistant to almost all classes of antibiotics with the possible exception of colistin. [76,77]
-
NDM-1-producing Enterobacterales has spread across Europe. In a survey conducted in 29 European countries, cases were reported in 13 countries. [76] Majority of the cases had a history of travel to the Indian subcontinent. Many countries have developed their own national guidelines to deal with the problem of NDM-1. [76]
-
The first NDM-1-producing E. coli in Hong Kong was isolated in October 2009 from a patient with urinary tract infection with travel history to India. [78] Several cases of IMP-4 were found in hospitalised patients since mid-2009 in Hong Kong. [79] The first KPC-2-producing K. pneumoniae was described in February 2011. [80]
-
The spread of NDM-1 is probably due to the huge selection pressure created by widespread non-prescription use of antibiotics in India [81] and involvement of promiscuous mobile elements in the gene’s dissemination. [82]
-
A local review of the NDM detected from 2009–2014 was performed by the CHP. From 2009–2013, there was a gradual rise of NDM cases detected, ranging from 1 to 19 patients, but there were no local cases of NDM detected during this period. Twelve local cases of NDM were first detected in 2014, where four patients had signs of infection. Majority of the imported NDM cases were from China, followed by India and other South East Asian countries. [83]
-
A local study in 2016 investigated the clonality and mechanism of resistance of 92 strains of CRE isolated between 2010 and 2012. Only 10% were genotypic CPE confirmed by polymerase chain reaction (PCR). Porin loss combined with AmpC and/or CTX-M type ESBL was the major mechanism of resistance of the CRE isolated. [84]
-
Plasmid-mediated colistin resistance by mcr-1, a gene that can be transferred horizontally among bacteria has been first described in China in both food animals and human. [85] Hong Kong has also detected CPE with mcr-1. [86] The coexistence of MCR-1 with carbapenemase (e.g. NDM, KPC) has been described in China, [87–89] South America, [90] Singapore, [91] and Germany. [92]
-
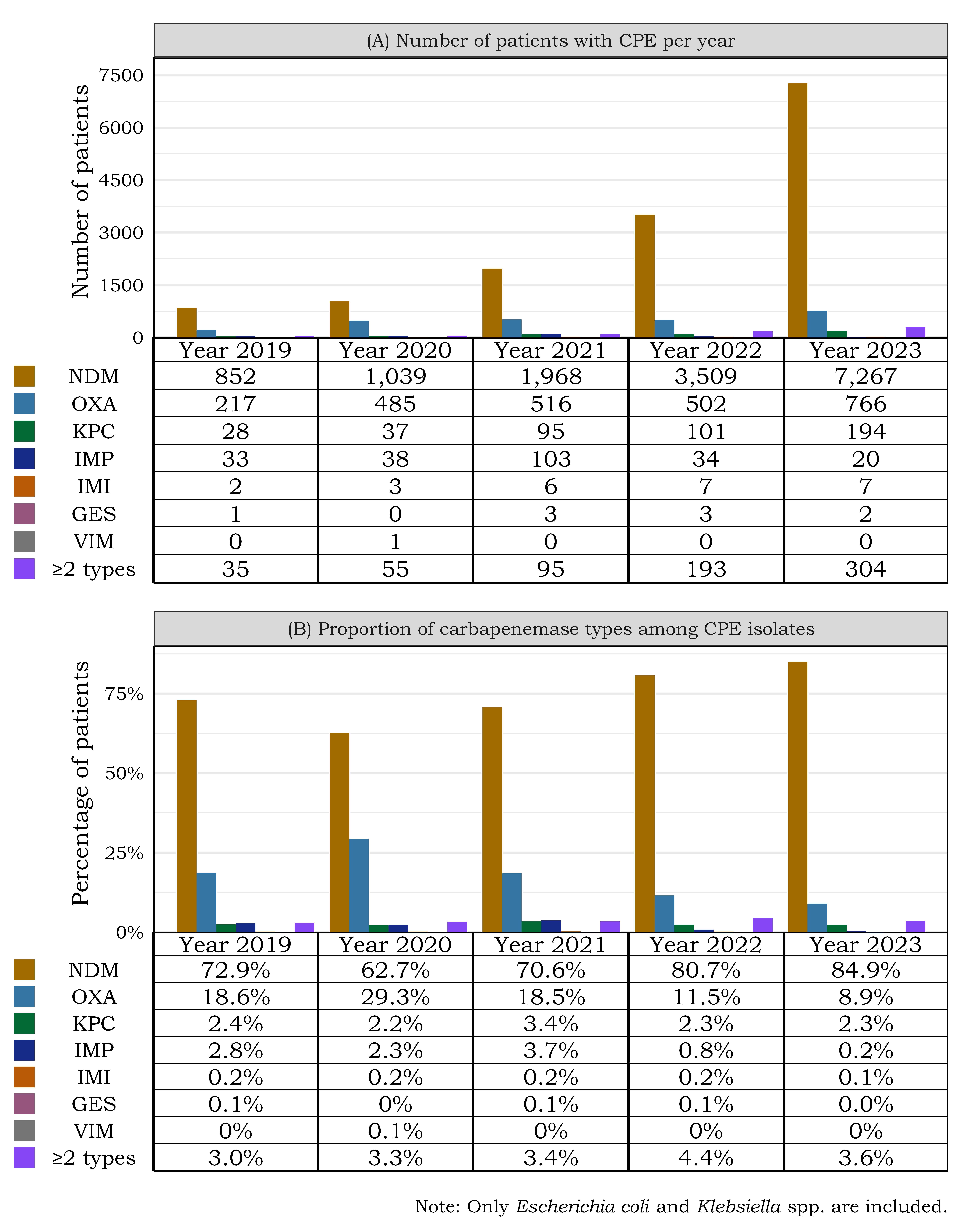
A local study on epidemiology and risk of CPE carriage in hospital showed an increasing trend from 2011 to 2019, with NDM as the predominant enzyme produced. [93,94] There is a significant rise of number of CPE isolates sent to the Public Health Laboratory Services Branch (PHLSB) for confirmation, from 1,868 in 2019 to 9,896 in 2023, where NDM accounts for 76.4% of all CPE, consistent with previous published findings (Figure 1.4).
-
CPE has been found in various food items, both raw and ready-to-eat, leading to potential silent and widespread dissemination within the community. Based on local surveillance data from the Centre for Food Safety, the detection rates of ESBL-E and CPE in raw chicken, pork, and beef were 74.3% and 29.4%, 71.9% and 32.0%, and 61.8% and 9.6%, respectively. [95] In ready-to-eat foods such as vegetable salad, sashimi, cut fruits, siu mei, and lo mei, 9% tested positive for ESBL-E, while 0.3% tested positive for CPE. [96]
|
Class A |
Metallo-β-lactamase |
OXA carbapenemase |
|---|---|---|---|
Molecular class |
Class A |
Class B |
Class D |
Functional class |
2f |
3 |
2d |
Gene location |
Usually transposon |
Usually plasmid |
Usually plasmid |
Examples |
KPC¹ |
IMP¹ |
OXA-23, 24, 51, 58 (types in Acinetobacter spp.)² |
Found in |
Enterobacterales |
Non-fermenters and Enterobacterales |
Non-fermenters and Enterobacterales |
Inhibited by |
Clavulanate and tazobactam |
EDTA |
No effective inhibitor |
Active site |
Serine |
Zinc ion |
Serine |
Carbapenem |
Hydrolysed |
Hydrolysed |
Hydrolysed |
Aztreonam |
Hydrolysed |
Not hydrolysed |
Not hydrolysed |
Early β-lactam |
Hydrolysed |
Hydrolysed |
Hydrolysed |
Extended spectrum cephalosporin |
Hydrolysed (except SME) |
Hydrolysed |
Hydrolysed poorly |
¹ Seen in Hong Kong |
|||
² Common in Hong Kong |
|||